 MIST
MIST MIST
MISTYanhui Hu, Arunachalam Vinayagam, Ankita Nand, Aram Comjean, Verena Chung, Tong Hao, Stephanie E Mohr, and Norbert Perrimon. 2017. “Molecular Interaction Search To\ ol (MIST): an integrated resource for mining gene and protein interaction data.” Nucleic Acids Res, 46, D1, Pp. D567-D574.
Details/ Paper \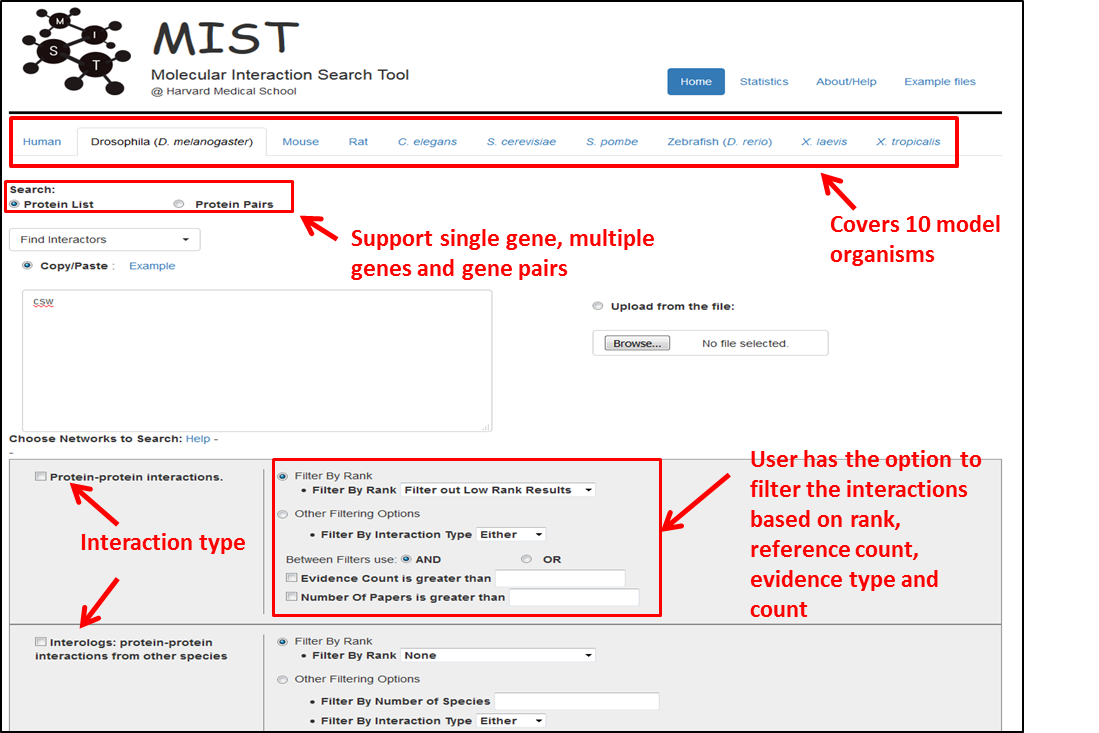
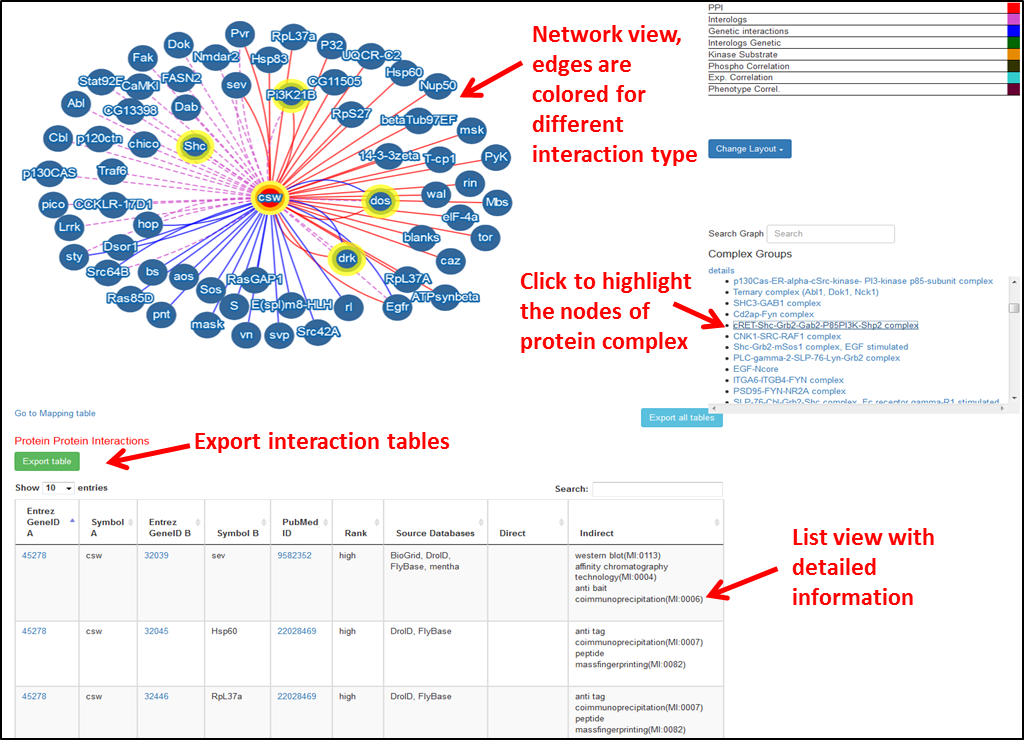
To download a network, right click on the network. An option to download a png/jpg or cytoscape js file will pop up. To download individual tables, click the “Export table” button on the top of each table. Alternatively, you can click the “Export all tables” button below the network to export all tables at once as .txt format.
Overview of possible results when you enter a gene or protein list
You input a given a gene symbol or ID (or a list of them), or pairs, and all genes or pairs have interactors in MIST. In this case, MIST will find the interactions and display them.
Example: For Human gene input- 81570,10994
For all or a few of your input gene symbols or IDs, no interactions are present in MIST, so no interactions are shown. The inputs will show up as green nodes without edges. These inputs will also be listed in the table “no interactions found”.
Example: For Drosophila gene input in ppi dataset- CG43295
If your input gene symbol or ID is misspelled or otherwise not in the database, then MIST will not find any matches. In this case, no interactions will be found, and you will see a red bar on the top saying following genes not found. If there is a mix of ‘good’ IDs and IDs that are not found, then results for the good IDs will be displayed, and the ‘bad’ IDs will be noted at the red bar on the top.
Example: In any case input - foo
If your input gene symbol or ID has been split into two genes in a more recent annotation of the genome (e.g. Gene A is now called Genes B and C), then both of the IDs that map to your input ID will be displayed (e.g. MIST displays results for B and C). However, if one of the newly annotated genes has the original ID, then MIST will only use this ID (e.g. Gene A is now called A and D, and MIST displays results for A).
Example: For Human gene input - A1B
The Molecular Interaction Search Tool (MIST) is a comprehensive resource of molecular interactions. MIST currently supports several species, including fly, mouse and human. With MIST, you can mine known physical interactions and infer interactions using other supportive evidence as well as similar genes by correlation analysis. The web interface allows users to retrieve interacting or similar genes in table format as well as visualize these interactions as networks.
“PPI” refers to physical interactions between two or more proteins.Both binary and complex-based PPI data are available. This information was compiled by integrating experimentally identified PPIs as annotated in BioGRID, IntAct, mentha, DIP, DroID, HPRD, FlyBase, PomBase.
“Interologs-ppi” are interactions predicted based on PPI data obtained for orthologous proteins. For example, a pair of Drosophila proteins would be considered interologs-ppi (i.e. they are predicted to interact) if the human orthologs of these proteins were found experimentally to interact with each other. We annotated interologs as follows. First, experimentally identified PPIs were compiled from major PPI databases. Next, the relevant proteins were mapped to specific species using ortholog annotation from DIOPT database. In case of one to many ortholog mapping, we selected the best ortholog gene using the DIOPT score as the indicator of highest-confidence ortholog relationships (Hu et al., 2011). In addition, we also filtered out any orthologous with low rank and/or score below 3.
Genetic interactions indicate that the effects of mutations in one gene can be modified by mutation of another gene. Genetic interactions were collected from BioGrid, IntAct, DroID and FlyBase. Interologs (genetic interaction): “Interologs-GI” are interactions predicted based on genetic interaction data obtained for orthologous proteins. Orthologous mapping for interolog-GI is the same as interolog-ppi (see above).
Kinase-substrate interactions are defined here as inferred interactions between a kinase and its potential substrate(s). We selected experimentally verified phosphorylation sites. Then we used the NetPhorest program to predict what kinase phosphorylated the site. This program is based on probabilistic sequence models of linear motifs to predict kinase-substrate relationship.
A phenotype correlation network was built based on gene pairs that share the similar or opposite phenotypes in cell-based functional genomic screens (Vinayagam et al., 2014). Very often, gene pairs that show significant phenotype correlations are part of same protein complex or involved in similar biological processes. A positive correlation suggests an activation-type relationship between the genes; a negative correlation suggests an inhibitory relationship between the genes.
This network was built on a data set created as follows (Sopko et al. 2014). First, specific kinases were disrupted in vivo. Next, changed phosphosites present in perturbed backgrounds vs control were identified. Then, a phospho-correlation network was built based on protein pairs that share similar or opposite pattern of phosphosites in kinase-deficient phosphorylation profiles. We have previously shown that such phospho correlation exist between functionally related proteins, e.g. they can be used to infer a kinase-substrate interaction (Sopko et al 2014). MIST contains phospho correlation relationships for Drosophila and S.cerevisiae.
We define expression correlation as the correlation of mRNA expression levels across many experimental conditions, cells and tissue types. MIST uses expression correlation information from DGET. Gene pairs with Pearson correlation coefficient >= 0.85 with at least 3 datasets are displayed.
| Source | Version or date of dowload | Interactions included in MIST (vs3) | Interactions included in MIST (vs4) | Increase | Non-overlapping interactions (vs3) | Non-overlapping interactions (vs4) | Interaction types | Species for MIST |
|---|---|---|---|---|---|---|---|---|
| DIP | dip20170205 | 106,660 | 106,660 | 0 | 6,517 | 921 | PPI | 10 |
| DroID (including DPiM) | v2018_8 | 247,816 | 280,617 | 13 | 129,219 | 128,632 | PPI,GI | 1 |
| BioGrid | vs3.5.171 | 1,827,231 | 2,114,921 | 16 | 1,105,722 | 1,198,044 | PPI,GI | 9 |
| IntAct (including MINT) | 5/23/2019 | 634,547 | 764,334 | 20 | 16,381 | 56,505 | PPI,GI | 10 |
| FlyBase | FB2019_2 | 64,007 | 78,661 | 23 | 21,192 | 2,881 | PPI,GI | 1 |
| HPRD | Release9 | 76,233 | 76,233 | 0 | 23,363 | 22,743 | PPI | 2 |
| PomBase | 6/22/2017 | 5,290 | 5,290 | 0 | 3,624 | 3,495 | PPI | 1 |
| mentha | 5/13/2019 | 1,020,351 | 1,202,463 | 18 | 9,454 | 12,108 | PPI | 9 |
| HumanMAPK | manuscript table (2010) | 4,530 | 4,530 | 0 | 2,941 | 2,911 | PPI | 1 |
| WormBase | WS270 | NA | 53,078 | NA | NA | 29,999 | PPI,GI | 1 |
| MIST (without interologs) | vs4 | 2,376,341 | 2,737,466 | 15 | NA | NA | PPI,GI | 10 |
| MIST (including interologs) | vs4 | 13,573,897 | 16,441,356 | 21 | NA | NA | PPI,GI | 10 |
information about downloads and linkouts is on the Downloads/Linkout" page.
" API infomration.
DRSC/TRiP Functional Genomics Resources
Department of Genetics
Harvard Medical School
New Research Building, Room 336
77 Avenue Louis Pasteur Boston, MA 02115
Email: perrimon@receptor.med.harvard.edu