@ Harvard Medical School
Intro Video:
About
InsulinNet provides access to a comprehensive Drosophila InR/PI3K/Akt network generated at Perrimon Lab (Harvard Medical School) by combining biochemical, genetic and computational approaches.
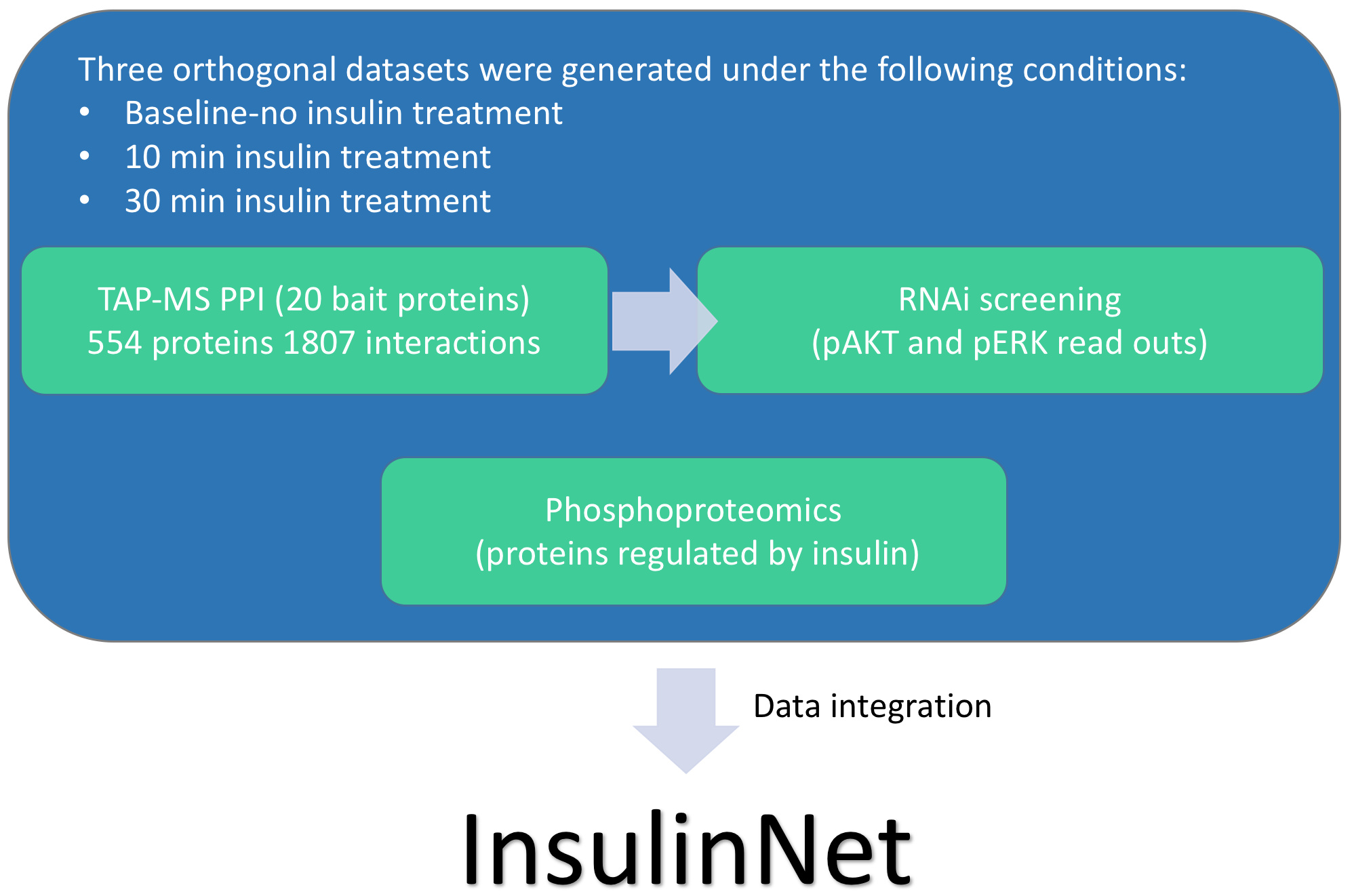
Building a comprehensive Drosophila insulin signaling network (InsulinNet)
The user interface was built using integrated data from protein-protein interaction (PPI), RNA interference (RNAi), and phosphoproteomics networks, together with cytoscape.js. The JSON files for these networks have a predefined layout and style, including predefined node color, shape, and edge colors. These were exported from Cytoscape and provided as an input to a Cytoscape javascript library in order to create the platform.
TopHow to use InsulinNet
The platform provides access to InsulinNet-PPI (PPI network generated using Tandem Affinity Purification followed by Mass Spectrometry), integrated InsulinNet (PPI network integrated with RNAi and phosphoproteomic data) and a protein complex view of InsulinNet. You can query the proteins in this network with the help of search box.
Search Box:
- Enter a FlyBase gene ID or gene symbol in the box (you will get suggestions after the 2nd letter you type for ID and symbol).
- Select and click on your gene of interest from the list.
- The layout on the left hand side will be refreshed, showing the specific sub-network associated with the gene you selected (query gene and its interacting partners).
Citing InsulinNet:
A. Vinayagam, M. Kulkarni, R. Sopko, X.Sun, Y. Hu, A. Nand, C. Villalta, A. Moghimi, X. Yang, S. E. Mohr, P. Hong, J. Asara, N. Perrimon, An integrative analysis of the InR/PI3K/Akt network identifies the dynamic response to Insulin signaling. (Manuscript in preparation) TopContact us
Perrimon lab Department of Genetics Harvard Medical School New Research Building, Room 336 77 Avenue Louis Pasteur Boston, MA 02115 Fax: (617)432-7688 Email: perrimon@receptor.med.harvard.eduTop